Abstract
Sickle Cell Disease (SCD) affects more than 14 million people globally, primarily in economically disadvantaged populations. Its management requires early diagnosis, monitoring, and treatment throughout the lifespan of the patient. Early diagnosis of SCD remains a challenge in the developing world due to requirements for sophisticated lab equipment and skilled personnel. American Society of Hematology with other organizations launched the Sickle Cell Disease Coalition, calling for improved patient care. In low-income countries, notably those in Sub-Saharan Africa, the mortality rate for children <5 years with SCD is 50-80%, without early diagnosis and basic care. Access to testing is lacking in these areas since existing SCD screening methods are expensive and resource intensive. The situation calls for an accurate and affordable diagnostic tool designed for point-of-care (POC). To address this need, we developed HemeChip, an accurate, affordable, and rapid "microchip electrophoresis" test, packaged in a small, easy-to-use, and low-cost mass-producible platform. HemeChip identifies SCD and other critical hemoglobinopathies as well as provides relative percentages of hemoglobin types. HemeChip is the first paper-based microchip electrophoresis platform that performs both qualitative and quantitative analysis of hemoglobins. Here, we build on our previous work (Blood 2015 126:3379) by presenting an initial clinical performance analysis of HemeChip's results, compared with High Performance Liquid Chromatography (HPLC), the clinical gold standard.
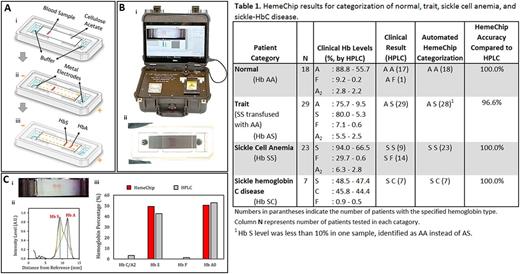
The compact single-use microchip of the HemeChip platform encompasses cellulose acetate paper, in which the hemoglobin separation takes place, as well as integrated stainless-steel electrodes (Fig. 1A). The HemeChip test is performed in four steps: (i) finger/heel prick equivalent amount of whole blood (diluted and lysed) is applied to the microchip (Fig. 1A); (ii) microchip is placed inside the HemeChip reader (Fig. 1B); (iii) real time images of the hemoglobin electrophoresis are captured during the test by the portable reader; and (iv) custom built-in software automatically analyses the results real-time, extracts relevant peak positions, and performs quantitative analysis (Fig. 1C) similar to HPLC. Total procedure takes less than 10 minutes.
HemeChip reliably detects hemoglobin types and categorizes patient samples as normal (Hb AA), sickle cell trait (Hb SA), sickle cell anemia (Hb SS), and sickle-hemoglobin C disease (Hb SC) (Table 1). All 77 blood samples tested were obtained under Institutional Review Board (IRB) approval. Tests were performed by both experienced and minimally-trained personnel. Test results for both categories did not introduce discrepancy or variation in accuracy. The HemeChip tests correctly categorized patient samples as Hb AA, Hb SA, Hb SS, and Hb SC with 96.6%-100% accuracy compared with HPLC (Table 1).
HemeChip provides high accuracy for hemoglobin phenotype detection and excellent compliance with the clinical gold standard for quantification of the results, which suggests that the HemeChip platform could address the unmet need for screening in regions where SCD is prevalent. Field-testing of HemeChip for SCD screening is ongoing in sub-Saharan Africa.
Acknowledgments: This work was supported by: Case-Coulter Translational Research Partnership, Clinical and Translational Science Collaborative of Cleveland (National Center for Advancing Translational Sciences), NIH Center for Accelerated Innovation at Cleveland Clinic, and Center for Integration of Medicine & Innovative Technology.
Figure 1: HemeChip overview and initial clinical testing results. (A) Illustration of hemoglobin separation during a HemeChip test. (i) initial, (ii) intermediate, and (iii) final phase of the separation in HemeChip. (B) (i) Portable HemeChip Reader with custom built-in analysis software, (ii) Ssingle use HemeChip cartridge microchip after a test. (C) HemeChip image analysis showing identification and quantification for an Hb SA blood sample. (i) Captured image of hemoglobin separation. (ii) Averaged pixel plot along the HemeChip length reveals intensity peaks and hemoglobin types. (iii) Baseline corrected area under curve determines the relative percentage of hemoglobins compared with HPLC.
Little: Hemex Health: Equity Ownership. Gurkan: Hemex Health: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal